Introduction to R
16. Comparing means
Independent samples t-tests are easy in R. Try:
t.test(bmi ~ gender, var.equal = TRUE, conf.level = 0.95, data=data)
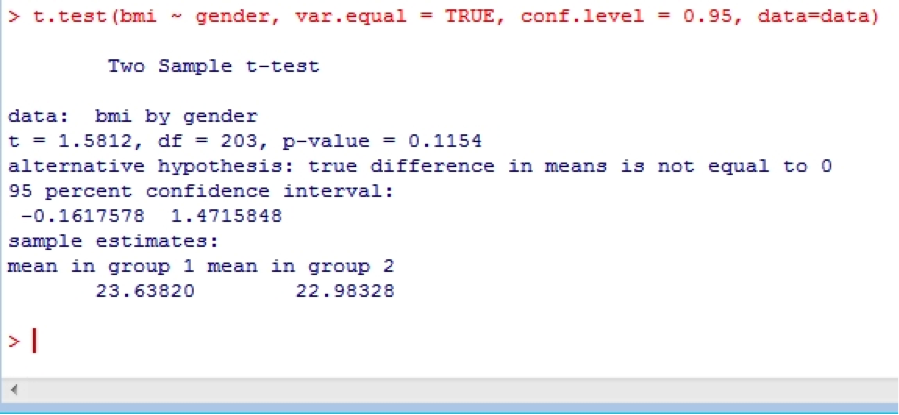
This uses the confidence level of 0.95 (conf.level = 0.95) which is the default, but you could use 0.99 if you like. Note that this test assumes equal variances (var.equal = TRUE), which is again the default, but you could change this to FALSE if need be. To test whether or not the variances are equal, use:
var.test(bmi ~ gender, data = data)
There is also the Fligner-Killeen test of homogeneity of variances, which a non-parametric test and apparently robust against departures from normality:
fligner.test(bmi ~ gender, data=data)
Oneway analysis of variance is relatively straight forwards. However, this is best done by creating an object for the results of the ANOVA, and requires that the grouping variable be a made into factor. For example for a comparison of BMI be boot comfort rating, we would use comfortf as it is a factor, and an object “anova_bmi_comf” to store the results:
anova_bmi_comf <- aov(bmi ~ comfortf, data = data)
anova_bmi_comf
summary(anova_bmi_comf)

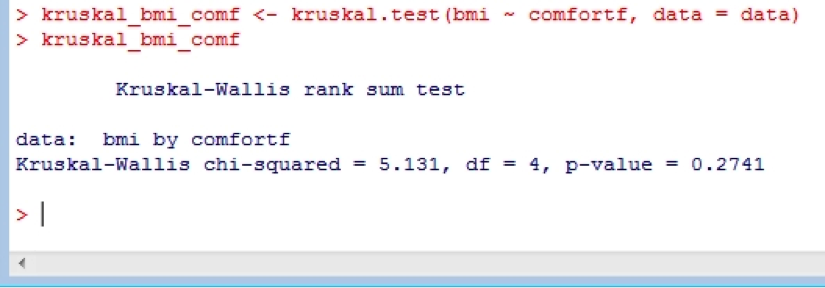
Non-parametric versions of the t-test and ANOVA are also available. For example:
kruskal_bmi_comf <- kruskal.test(bmi ~ comfortf, data = data)
kruskal_bmi_comf