INTRODUCTION
Much of the information found in this chapter is adapted from the literature of the manufacturers of the kit systems described. Where possible, manufacturers details will be listed.
The brand used in practicals will vary from time to time, and so details of exact requirements/procedures may not be the same. Always check the instructions supplied with the kit.
MICROBACT™ 24E GRAM-NEGATIVE IDENTIFICATION SYSTEM
Supplier: Oxoid Australia
INTENDED USE
The Microbact™ Gram-negative system is to be used for the identification of aerobic and facultatively anaerobic Gram-negative bacteria (Enterobacteriaceae and miscellaneous Gram- negative bacteria)
These kits come in a number of configurations. you will need an "A" strip and a "B" strip to make up a 24E test.
PRINCIPLE OF THE TEST
The Microbact™ Gram-negative system is a standardised miniaturised system which is designed to replace conventional biochemical tests in test tubes and other media. Reagents are suppled dried into wells in a microtitre tray; when a culture suspension in saline is added, the reagents are rehydrated; the tray is incubated and specific colour changes occur depending on the identity of the organism. Colour changes occur due to pH changes and various substrate utilisations. Refer to the table of reactions below for the substrates contained in each well, the specific reaction principle, and colour changes.
SET UP PROCEDURE
Isolation
- Pure cultures grown overnight should be used for testing. Non-selective media is preferred.
- Before use, perform an oxidase test on the organism to be identified and record the result on the result sheet. The oxidase result is important for determining which database to access.
Preparation of inoculum
Pick 1-3 isolated colonies from an 18-24 hour culture and emulsify in 5.0ml of sterile saline. Mix thoroughly to prepare a homogeneous suspension.
Inoculation
- Prepare the tray - place in the holding tray and label the plastic covering on the tray with a texta. Peel back to expose the 2 rows which constitute one test.
- Using a sterile Pasteur pipette add 4 drops (approximately 100µl) of the bacterial suspension, or half fill each well in the set.
- Overlay the substrates underlined (in black) on the holding tray with sterile mineral oil, i.e. wells 1, 2 , 3, 20 and 24 for 24E. (Well 20 for 24E is not overlayed with oil for oxidase- positive, miscellaneous Gram-negative bacilli.)
- Replace the peel back covering to seal the wells and incubate the tray at 37oC aerobically overnight.
- To determine the purity of the inoculum, it is advisable to inoculate a solid non-selective medium (e.g. nutrient agar) with the test suspension to act as a culture purity check.
Reading the test strip
The 24E strip is read at 24 hours when identifying Enterobacteriaceae. All systems should be read after 48 hours for the identification of Miscellaneous Gram-negative bacilli.
Remove the strips or tray from the incubator, peel back the sealing tape.
Add the following reagents (wells which must have reagents added have green circles around them):
- Well 8 (Indole production) - add 2 drops of Indole (Kovacs) reagent. Evaluate within 2 minutes of the addition of the reagent.
- Well 10 (Voges-Proskaüer reaction) - add 1 drop each of VPI reagent and VPII reagent. Evaluate 15 to 30 minutes after the addition of reagents.
- Well 12 (Tryptophan Deaminase) - add 1 drop of TDA reagent (FeCl3). Test can be evaluated immediately after the addition of the reagent.
Record all positive results. The reactions are evaluated as positive or negative by comparing them with the colour chart. Record the results under the appropriate heading on the report form. For aid in interpreting reactions, refer to the Table of Reactions.
Note:
- The gelatin well (well 13 for 24E) must be read at 24-48 hours for Enterobacteriaceae and at 48 hours for miscellaneous Gram-negative bacilli (MGNB). Hydrolysis of gelatin is indicated by dispersal of the black particles throughout the well.
- The arginine reaction (well 24 of 24E) is interpreted differently at 24 hours and 48 hours incubation.
24 Hours Incubation (Enterobacteriaceae):
- Yellow - Negative
- Green-blue - Positive
48 Hours Incubation (MGNB):
- Yellow-green - Negative
- Blue – Positive
Additional Test
Nitrate Reduction Test
This test is performed in well 7 (ONPG) AFTER reading the ONPG reaction. One drop of Nitrate reagent A and 1 drop of Nitrate reagent B is added to the well. Production of a red colour within a few minutes of the addition of the reagent indicates that nitrate reduction to nitrite (NO2) has occurred. A small amount of zinc powder should be added to those wells which exhibit a yellow colour after the addition of the nitrate reagents. This will determine whether nitrate has been reduced completely to nitrogen gas (N2). The results should be interpreted as follows:
After the addition of Nitrate reagents A and B:
- Red colour - NO2 Positive
- Yellow colour - NO2 Negative
On the addition of zinc powder:
- Yellow colour - Positive (N2)
- Red colour - Negative (N2)
All organisms belonging to the family Enterobacteriaceae reduce nitrates to nitrites and give a positive reaction.
Table of Reactions
| Well Number | Designation | Reaction Principle | Negative | Positive | Comments |
| 1 | lysine | lysine decarboxylation | yellow | blue-green | Bromothymol blue indicates formation of specific amine - cadaverine |
| 2 | ornithine | ornithine decarboxylation | yellow-green | blue | Green should be regarded as a negative reaction. The pH shift indicated by bromothymol blue caused by formation of the specific amine putrescine is greater than that caused by lysine decarboxylation. |
| 3 | H2S | production of H2S | straw coloured | black | H2S is produced from thiosulphate. H2S reacts with ferric salts in the medium to form a black precipitate. |
| 4 | Glucose | glucose fermentation | blue-green | yellow | Bromothymol blue indicator changes from blue to yellow when the carbohydrate is utilised to form acid. |
| 5 | Mannitol | mannitol fermentation | blue-green | yellow | Bromothymol blue indicator changes from blue to yellow when the carbohydrate is utilised to form acid. |
| 6 | Xylose | xylose fermentation | blue-green | yellow | Bromothymol blue indicator changes from blue to yellow when the carbohydrate is utilised to form acid. |
| 7 | ONPG | Hydrolysis of o-nitrophenyl-β-d-galactopyranoside (ONPG)by action of β-galactosidase |
colourless | yellow | β-galactosidase hydrolysis of the colourless ONPG releases yellow orth-onitrophenol. |
| 8 | Indole | formation of indole from tryptophan | colourless | pink/red | Must add Kovac's reagent which forms a pink/red complex with indole. |
| 9 | Urease | hydrolysis of urea | colourless | pink | as urea is hydrolysed, it releases ammonia, causing the colour of the phenol red pH indicator to change to pink. |
| 10 | VP | acetoin production (Voges-Proskauer reaction) | colourless | pink | Must add VP reagents A (α-naphthol) and B (creatine). These react with acetoin (produced from glucose) to form a pink complex. |
| 11 | Citrate | Utilization of citrate as sole carbon source | green | blue | Citrate is the sole carbon source, which if utilized results in a pH rise, indicated by bromothymol blue. |
| 12 | TDA | deamination of tryptophan to produce indolepyruvate | colourless | dark brown/black | Must add FeCl3. Deamination of tryptophan produces indolepyruvate which reacts with ferric ions to turn brown/black. |
| 13 | Gelatin | gelatin liquefaction | colourless | uniformly black | Liquefaction of the reagent releases particles of black pigment diffusely throughout the well. |
| 14 | Malonate | Malonate inhibition of the conversion of succinate to fumarate | green | blue | Indicated by change in bromothymol blue pH indicator. |
| 15 | Inositol | fermentation of the sugar | blue-green | yellow | Bromothymol blue indicator changes from blue to yellow when the carbohydrate is utilised to form acid. |
| 16 | Sorbitol | fermentation of the sugar | blue-green | yellow | Bromothymol blue indicator changes from blue to yellow when the carbohydrate is utilised to form acid. |
| 17 | Rhamnose | fermentation of the sugar | blue-green | yellow | Bromothymol blue indicator changes from blue to yellow when the carbohydrate is utilised to form acid. |
| 18 | Sucrose | fermentation of the sugar | blue-green | yellow | Bromothymol blue indicator changes from blue to yellow when the carbohydrate is utilised to form acid. |
| 19 | Lactose | fermentation of the sugar | blue-green | yellow | Bromothymol blue indicator changes from blue to yellow when the carbohydrate is utilised to form acid. |
| 20 | Arabinose | fermentation of the sugar | blue-green | yellow | Bromothymol blue indicator changes from blue to yellow when the carbohydrate is utilised to form acid. |
| 21 | Adonitol | fermentation of the sugar | blue-green | yellow | Bromothymol blue indicator changes from blue to yellow when the carbohydrate is utilised to form acid. |
| 22 | Raffinose | fermentation of the sugar | blue-green | yellow | Bromothymol blue indicator changes from blue to yellow when the carbohydrate is utilised to form acid. |
| 23 | Salicin | fermentation of the sugar | blue-green | yellow | Bromothymol blue indicator changes from blue to yellow when the carbohydrate is utilised to form acid. |
| 24 | Arginine | presence of arginine dehydrolase | 24 hours - yellow
48 hours - yellow/green |
24 hours - green/blue
48 hours - blue |
Removal of ammonia from arginine results in rise in pH indicated by bromothymol blue indicator. |
Interpretation
An octal coding system has been adopted for Microbact™. Each group of three reactions produces a single digit of the code. Using the results obtained, the indices of the positive reactions are circled. The sum of these indices in each group of three reactions forms the code number. This code is entered into the computer package. An example is shown below. Note that the first group of three reactions (oxidase, motility and nitrate) are not on the Microbact tray and must be performed separately. They are not included for an oxidase negative organism.
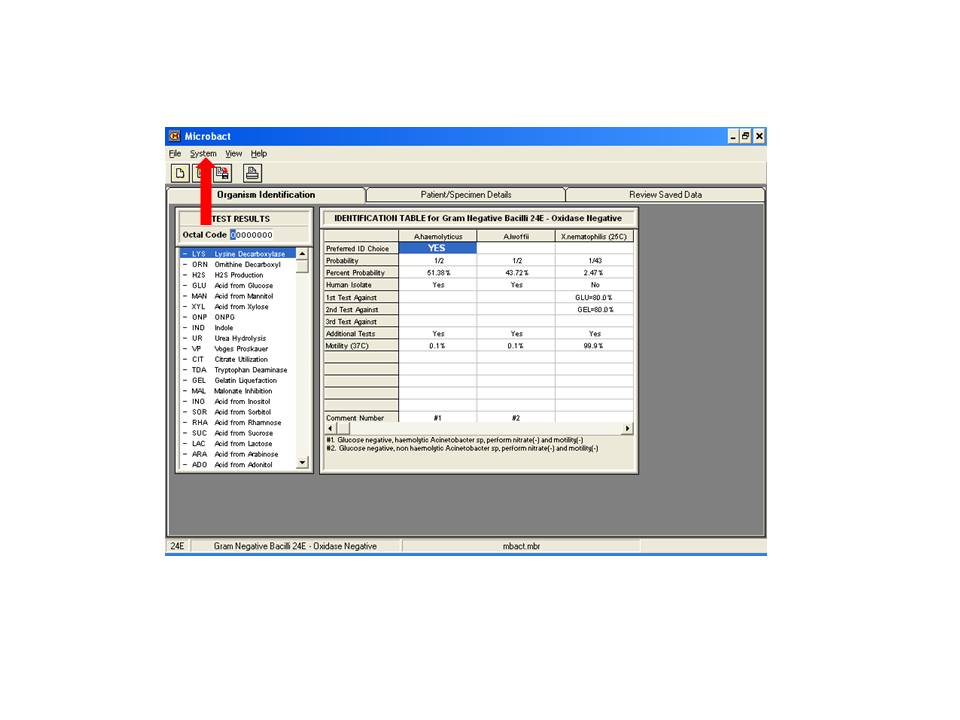
The Microbact™ Computer Aided Identification Package should be consulted for the identification choices. The database chosen depends on the result for the oxidase reaction. Computer aided identification Package – available on laboratory computers. When the Microbact program has loaded the page will look look this:
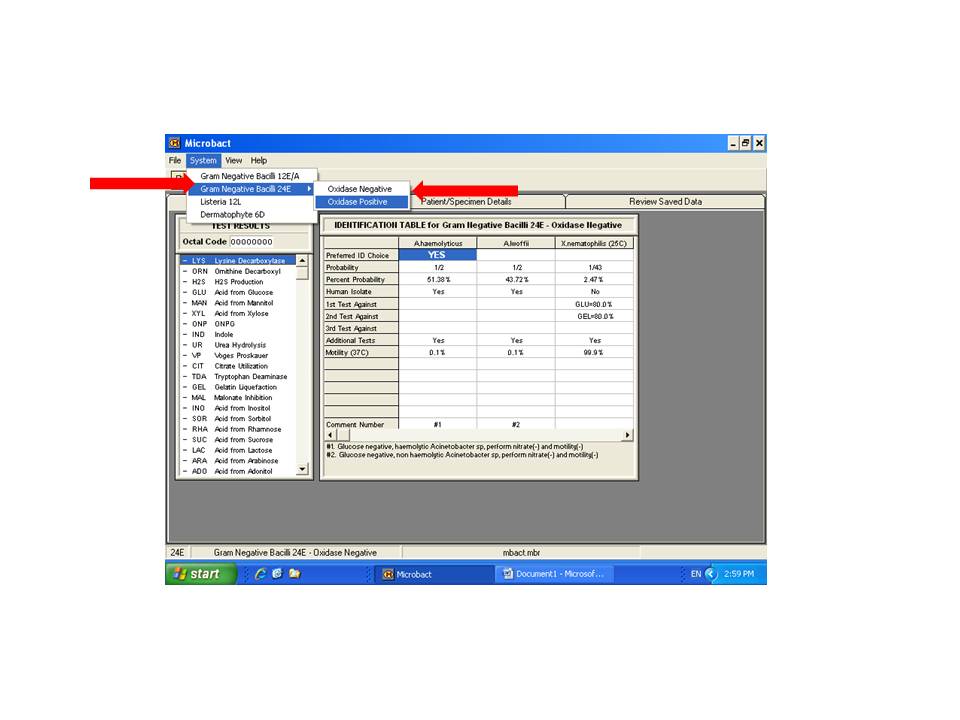
Click on "system" to select appropriate database (red arrow).
Select Gram Negative Bacilli 24E and appropriate oxidase reaction (negative for E. coli).
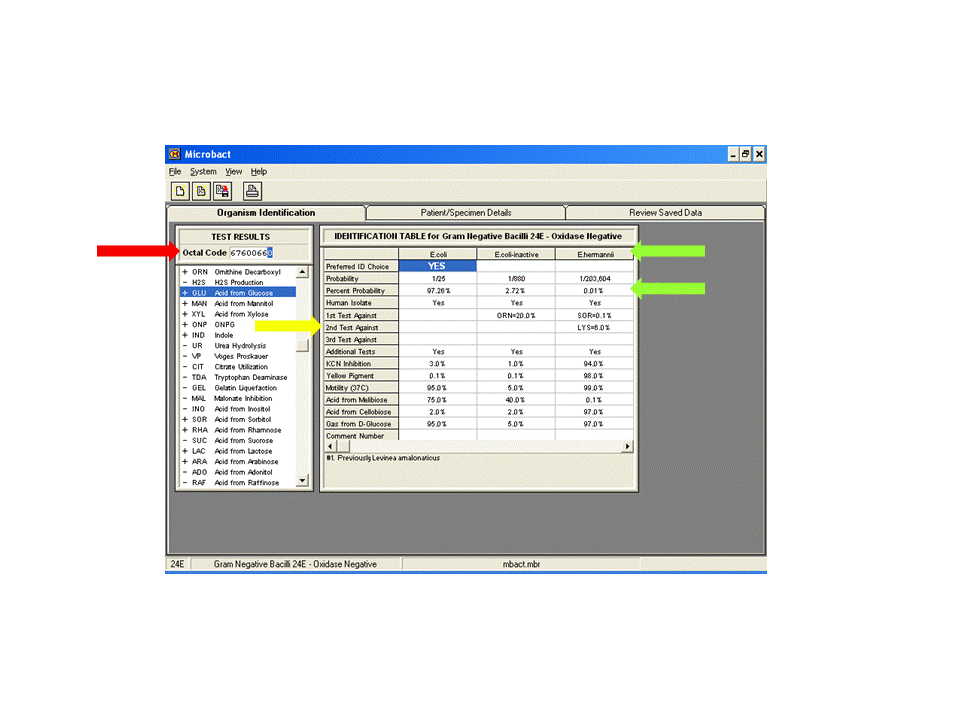
Type in the octal code (red arrow). The result will be displayed with percent probability (green arrows). If low percent, unusual or unexpected results occur, check reactions of 1st, 2nd and 3rd tests against (yellow arrow). These results may need to be repeated.
The percentage figure shown against the organism name is the percentage share of the probability for that organism as a part of the total probabilities for all choices. The three choices displayed may not necessarily total 100%.
Note: Miscellaneous Gram-negative bacilli - Weakly positive reactions are recorded as negative results. The results of tests for oxidase, nitrate reduction and motility are included as part of the reaction pattern. Using the results obtained, from each group of three reactions a 9 (nine) digit code number is produced.
Limitations
1. Some bacterial strains may have atypical biochemical reactions due to unusual nutritional requirements or mutations and may be difficult to identify.
2. Reactions obtained using the Microbact System may differ from published results using other substrate formulations. Prolonged incubation, insufficient incubation, improper filling of wells, or inadequate inoculum may lead to false results.
3. Species with low frequency of occurrence require additional testing.
4. Acinetobacter calcoaceticus var. anitratus will include those strains that have been designated as A. calcoaceticus, A. baumannii, and unnamed genospecies 3; most clinical isolates that are glucose-positive and nonhemolytic are A. baumannii.
5. The interpretation of mathematically calculated identification results requires trained personnel who should use judgement and knowledge in conjunction with the following information before accepting the ID of an organism: Gram-stain, colonial morphology, source of isolate, percent probability (degree of separation), tests against, additional test indications and results, frequency of ID choice and antibioGram.
6. A Gram-stain and oxidase test should be performed prior to set-up of tests. In addition, motility and nitrate test should be performed for miscellaneous Gram-negative bacilli.